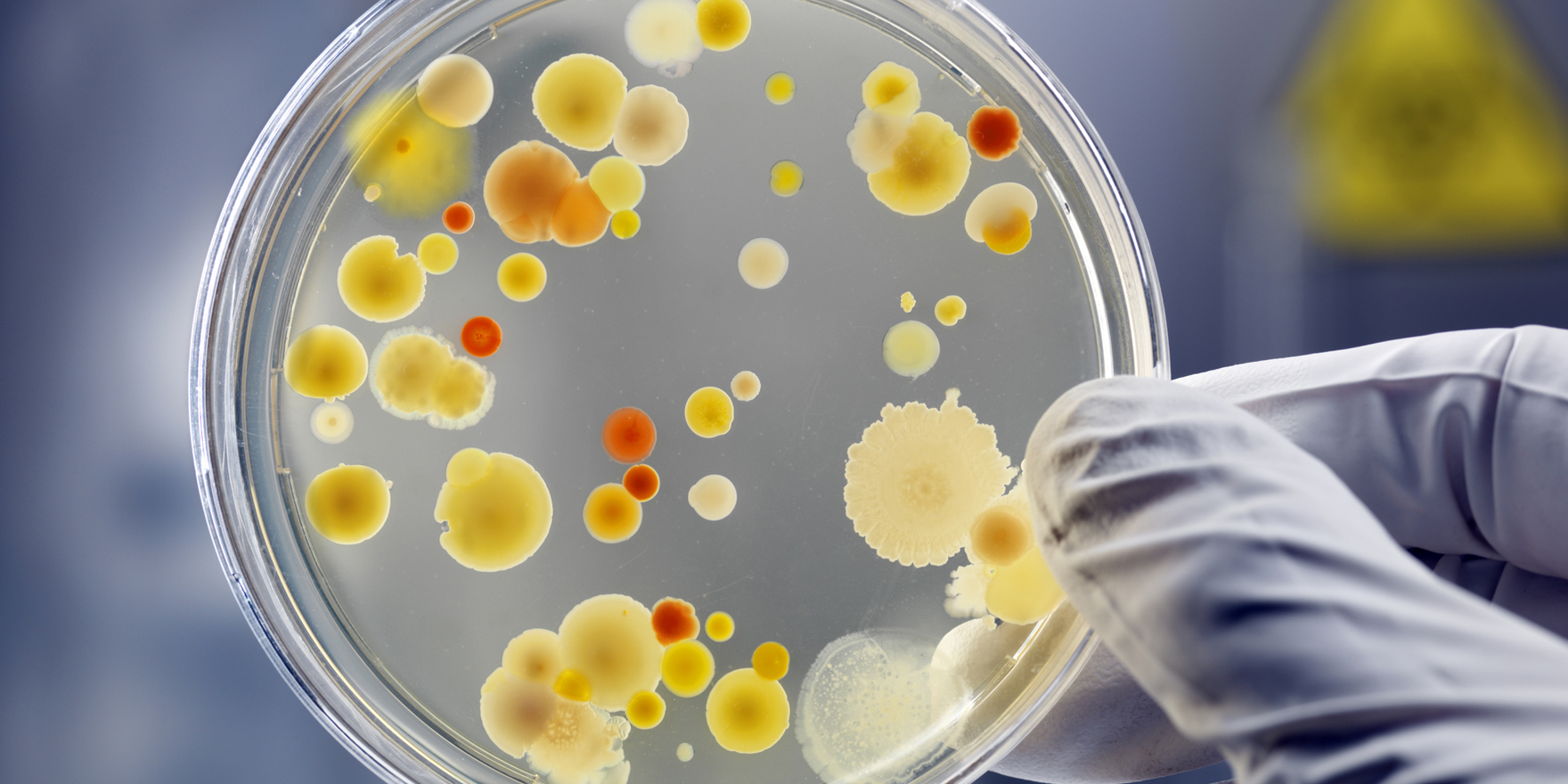
Imagine a future where a GP could quickly and effectively distinguish between bacterial and viral infections in their surgery
A new form of mass spectrometry promises to identify antibiotic resistant strains of bacteria and provide clinically actionable results in under a minute.
MALDI mass spectrometry has been used to identify bacteria and fungi since the 1990s, but the new technology, called REIMS, dramatically cuts the processing time.
Remarkably, REIMS can identify antibiotic resistant bacteria with around 87% accuracy, something which the existing technology cannot do.
Speaking at Pathology Update 2018 in Sydney, REIMS inventor Professor Zoltan Takats said the technology could prove quite relevant for infections such as septicaemia.
“It is actually much better than making a wild guess,” he said.
REIMS has never been used in a clinical setting, but the plans are under way to commercialise the technology through a university spinoff company.
Certifying the technology as a medical diagnostic tool was challenging, so his company was focusing first on launching a consumer healthcare product, Professor Takats, who is a professor of analytical chemistry at the Imperial College London, said.
REIMS makes efficiency gains by doing away with sample processing altogether, and by fully automating the system.
MALDI is a slow process that requires a person to scrape the bacteria culture off a petri dish with a toothpick, mix it with a matrix solution, dry the sample, and then hit it with UV light, which ionises molecules through a photochemical process.
REIMS skips all these steps by ionising bacterial cultures directly using an infrared laser, and then identifying the bacteria subspecies by detecting unique molecular markers.
Where REIMS get really futuristic, however, is its ability to detect bacteria in tissue samples, stool samples, histological slides and blood samples directly, something that has previously been impossible.
Using this method, a laboratory could analyse a stool sample without any sample preparation at all. This was a welcome development among lab personnel, who tended to regard working with messy stool samples as some kind of punishment, Professor Takats said.
When using human tissue or blood, REIMS could only correctly identify bacteria down to the genus, because there was too much noise from human molecular markers, he said. But using bacterial cultures, REIMS could correctly classify bacteria to subspecies level around 97% of the time.
While the process of taking inventions to market is a difficult one, Professor Takats imagines a not-too-distant future where a GP can use mass spectrometry in their practice to quickly distinguish between bacterial and viral respiratory infections using a nasal swab.
“The entire procedure, the swab and the analysis, can be done in a single minute, especially if the mass spectrometer is in the doctor’s office, which is not always the case but we hope that in five or 10 years it will be,” he said.
REIMS can also distinguish between different types of human tissue in situ and in real time. This feature formed the basis for another of Professor Takats’ inventions, the intelligent knife, which was unveiled in 2013.
The iKnife uses mass spectrometry to tell surgeons whether or not human tissue is cancerous based on its chemical signature. This helps surgeons preserve healthy tissue without missing any tumourous cells.
Professor Takats has also patented a second mass spectrometry technology, called DESI, which can profile bacterial communities. Mass spectrometry has never been used for this purpose before with researchers usually rely on DNA sequencing to investigate the microbiome, a process which can take a number of days.